|
Malaysia is home to a rich assemblage of parasitic flowering plants representing seven families, 46 genera and approximately 100 species. These plants are seldom considered candidates for conservation efforts, however, many species are important components of the tropical ecosystem that show complex associations with other organisms and unique biochemical features. Results of a phylogenetic analysis of 29 members of Santalales using a combined dataset of nuclear-encoded 18S rDNA and plastid-encoded rbcL sequences are presented. Sequences from representatives of three nonphotosynthetic, holoparasitic families often allied with Santalales, Balanophoraceae, Hydnoraceae and Rafflesiaceae, have been obtained from nuclear-encoded 18S rDNA and plastid-encoded 16S rDNA. As with 18S rDNA, the 16S rDNA sequences from all three holoparasite families showed an increase in the number of substitutions. The greatest increases were seen in Mitrastema and Hydnora, greater than values obtained from pairwise comparisons involving taxa as phylogenetically distant as angiosperms and liverworts. A case is made that these plants represent unique natural genetic experiments that offer a wealth of opportunity for molecular genetic and phylogenetic analyses.
When considering the reasons for conservation of biodiversity, one inevitably concludes that all involve value judgments that are, in essence, anthropocentric. For example, the value of a plant species might be determined by looking at its potential to provide food, drugs, natural products, or a new ornamental. In addition to this "commodity value," Norton (1988) also mentions amenity value, i.e. the value of enriching human experience. Given this, it may seem ironic or paradoxical to argue for the conservation of a parasite. The usual notion conjured up by the word is an organism that causes damage to its host, hence to include parasitic plants in conservation discussion may meet with some resistance or skepticism. Of course there are parasitic flowering plants that do damage their host plants (Striga, Orobanche, Arceuthobium, etc.), however, the vast majority of species are essentially benign causing little or no measurable effect on host fitness. So, the concept of parasite and pathogen are discrete - all pathogens are parasites but not the other way around.
One example of the conservation of a parasitic plant because of its commodity value can be seen with the European mistletoe. Viscum album is the source of biologically active molecules (Iscador) that are being used to treat cancer patients. Overcollecting, especially on hosts thought to confer desirable properties to the mistletoe, has resulted in a marked decline of this species in some areas prompting efforts at artificial inoculation and cultivation (Grazi 1984). There are numerous examples of rare mistletoes and at least one, Trilepidea adamsii of New Zealand, is now extinct owing to a combination of natural and anthropocentric factors (Norton 1991).
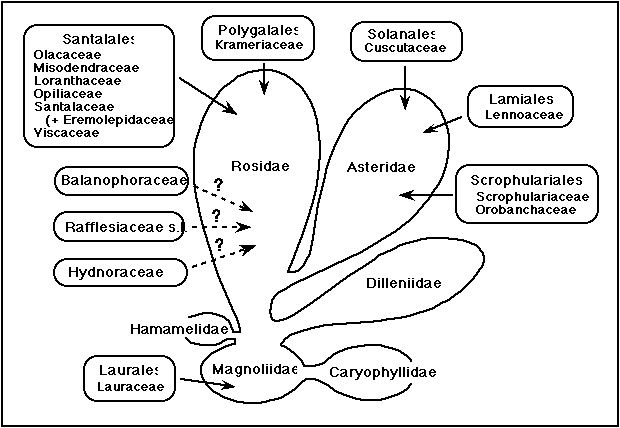
The number of angiosperm families that contain parasitic members varies depending upon the taxonomic system, but it appears that parasitism arose independently in flowering plants at least seven times and more likely nine times (Figure 1). Parasites range from chlorophyllous, facultative hemiparasites (Olacaceae, Opiliaceae, Krameriaceae) to obligate hemiparasites (Viscaceae, Loranthaceae) to nonphotosynthetic holoparasites (Balanophoraceae, Hydnoraceae, Lennoaceae, Rafflesiaceae, etc.). These plants show an amazing variety of specializations, some unique in the flowering plants (Kuijt 1969). These specializations involve features of morphology, anatomy, physiology, and reproductive biology. More recently, insight has been gained as to the organization of the nuclear and chloroplast genomes in these plants (see below).
Although approximately 3000 species of angiosperms have evolved parasitic associations, less than one tenth of these have become holoparasites, i.e. have lost chlorophyll and therefore photosynthetic function. These plants represent the most extreme manifestation of the parasitic mode, being totally dependent on their host for water, nutrients, and photosynthates. They must attach to both xylem and phloem and do so by means of highly specialized haustoria. Holoparasitism has evolved at least five times in angiosperms and is seen in Scrophulariaceae, Orobanchaceae, Lennoaceae and Cuscutaceae as well as three santalalean families - Balanophoraceae, Rafflesiaceae, and Hydnoraceae.
The purpose of this paper is to present an overview of some of our recent work with molecular evolutionary studies of parasitic plants, specifically those found in Malaysia. The groups we have focused on include all members of the order Santalales as well as Balanophoraceae, Hydnoraceae, and Rafflesiaceae. I will refer to this group as the "santalalean holoparasites" even though the affinities of the latter three families with this order are by no means clear. I will show that there exists in these plants poorly understood processes occurring at the molecular level that have not been previously documented in any other organism. Furthermore, these processes represent areas of active research that have significance beyond parasitic plants themselves. These plants therefore represent unique models suitable for a multitude of tests to further expand our knowledge of the evolutionary process. For these reasons, they are highly deserving of conservation efforts.
Malaysia is home to some of the richest biological diversity on earth, hence it is not surprising that a diverse assemblage of parasitic plants can also be found here. A compilation of the Malaysian representatives of Santalales and Rafflesiales (sensu Cronquist 1988) is shown in Table 1. This compilation, including peninsular Malaysia as well as Borneo, contains seven families, 46 genera and approximately 100 species. Many of the genera of Olacaceae are monotypic and four of these occur in Malaysia (Erythropalum, Harmandia, Ochanostachys, and Scorodocarpus). Similarly, monotypic members of Opiliaceae such as Champereia, Lepionurus and Melientha also occur here. Among the 74 genera of Loranthaceae, 14 occur in Malaysia. Five of the nine species of Lepeostegeres and the monotypic genus Lampas elmeri are endemic to Borneo. Seven genera of Santalaceae are known from Malaysia, however, the group requires further taxonomic work especially as regards generic boundaries among Cladomyza, Dendromyza, Dendrotrophe and Dufrenoya. The monotypic member of Balanophoraceae, Exorhopala ruficeps (Ridl.) Steen., is endemic to the Malay peninsula whereas Rhopalocnemis is monospecific (R. phalloides Jungh.) and is found throughout Indomalaysia.
Some of the most unusual and spectacular parasitic plants occur in the family Rafflesiaceae. All are extremely specialized, obligate holoparasites, often rare with very limited overall distributions. Two genera, Rafflesia and Rhizanthes, are large-flowered parasites of Tetrastigma (Vitaceae) and the third, Mitrastema (= Mitrastemon, sometimes placed in its own family Mitrastemonaceae) is a diminutive parasite of Fagaceae. The latter genus contains only two widely disjunct species: Mitrastema yamamotoi from Borneo, Sumatra, Indochina, and Japan and M. matudae from Mexico and Central America. Rhizanthes lowi is reported from Sumatra, the Malay Peninsula and Borneo and R. zippelii occurs in those regions plus Sumatra (Meijer and Veldkamp 1988). Members of the genus Rafflesia hold the honor of possessing the largest flower in the world. As shown in Table 2, eight species of Rafflesia occur in Malaysia. The taxonomy of the genus is still in flux, but there appears to be ca. 14 taxa, one of which are now likely extinct. Given that most species are naturally rare and adapted to stable, undisturbed forest environments, they are especially vulnerable to extirpation due to habitat loss (Meijer 1985, Ismail 1988).
Our research over the past six years has involved molecular phylogenetic relationships in Santalales. The first paper on this topic (Nickrent and Franchina 1990) used direct sequencing of nuclear rRNA and included results from only four members of this order. Since then, we have generated sequences from PCR products and, at present, have over 100 complete 18S rDNA sequences for this order. Relationships among 15 representative taxa were reported in Nickrent and Soltis (1995). To increase resolution of relationships, sequences of the chloroplast gene rbcL from 29 santalalean taxa plus five outgroup species were obtained. Eleven of these genera occur in Malaysia (compare with Table 1). Sequences of nuclear 18S rDNA (ca. 1800 bp) and rbcL (ca. 1430 bp) were analyzed using parsimony (minimum length Fitch trees) implemented by PAUP (Swofford 1993). Ten shortest trees of length 2262 were obtained and the majority rule consensus tree is shown in Figure 2. This molecular phylogenetic analysis confirms and expands upon previous concepts about interfamilial relationships in the order as explained below.
In agreement with Kuijt (1969), Olacaceae is basal in the order, but is composed of a very problematic assemblage of autotrophic and hemiparasitic woody plants. It is probably at least paraphyletic and likely polyphyletic. Additional sampling (e.g. Anacolosa, Erythropalum, Harmandia) is needed to fully circumscribe the phylogenetic limits of this family. A clade comprising Loranthaceae and Misodendraceae emerges next in the order. Loranthaceae are known to have diverged at the time Gondwana broke apart into South America and Africa (Barlow 1983), hence a number of biogeographic hypotheses are apparent and available for testing. When the 18S rDNA sequences for 22 genera of Loranthaceae are analyzed separately (results not shown), a notable feature of the phylogenetic tree is a dichotomy between the Old and New World genera. Both 18S and rbcL show a surprising relationship between the root parasite Schoepfia (Olacaceae) and the Loranthaceae. Schoepfia is also allied with a very unusual mistletoe, Misodendron, that is notable in being a stem parasite of southern hemisphere beech trees (Nothofagus) with wind-dispersed seeds. The relationship between Schoepfia and Loranthaceae is not so surprising, however, when one considers that three genera considered primitive in the family are also root parasites (Nuytsia, Atkinsonia, and Gaiadendron).
The next group to be derived in the order is the monophyletic family Opiliaceae that is composed of root parasitic shrubs, trees, and even lianas. Their close association from molecular data occurs despite being collected from Australia (Cansjera and Opilia), Costa Rica (Agonandra) and Malaysia (Champereia). Sampling within Santalaceae is currently limited to 11 genera, of which only two (Osyris and Santalum) have been sequenced for both 18S rDNA and rbcL. This family appears as a monophyletic group, but only after the inclusion of rbcL data. One surprising result is that two genera of the family Eremolepidaceae (Antidaphne and Eubrachion) are included within this clade. These two genera, along with Lepidoceras, were placed in their own family by Kuijt (1988), intermediate between Viscaceae and Loranthaceae by Bhatnagar and Vohra (1983), or as aerial members of Santalaceae by Wiens and Barlow (1971). The latter placement appears most in agreement with molecular data. The most derived family within the Santalaceae is the Viscaceae. Preliminary results from 18S rDNA sequencing indicates that Dufrenoya and Dendrotrophe (Santalaceae) are the closest relatives to Viscaceae within Santalaceae. This supports the notion that Viscaceae evolved in the Old World from aerially parasitic Santalaceae. Additional molecular work with the generic segregates of Henslowia (e.g. Cladomyza, Dendromyza, Dufrenoya) is needed to better understand the evolution of aerial parasitism that is found in all Viscaceae. It is apparent from viewing the cladistic tree that branches leading to some genera of Viscaceae (e.g. Arceuthobium) are longer than those leading to other genera in the order. This "stepped up" rate of evolutionary change has been the subject of much of our recent work.
The first indication that substitutions at nuclear 18S rRNA genes in plants are not behaving in a clocklike fashion was by Nickrent and Franchina (1990) where two members of Viscaceae showed significantly higher numbers of transitions than 12 other plants included in the study. All Viscaceae are obligate hemiparasites, however, the most dramatic demonstration of rate heterogeneity was seen when holoparasitic plants were examined (Nickrent and Starr 1994). The three santalalean holoparasite lineages, Balanophoraceae (represented by Balanophora), Hydnoraceae (Prosopanche) and Rafflesiaceae (Rafflesia and Rhizanthes) all show highly significant increases in the number of substitutions per site as compared with autotrophic outgroup taxa. The mean number of substitutions per site for the holoparasites was, on average, 3.5 times higher than autotrophs. Despite this high number of substitutions, the rRNA molecules retain secondary structure and are therefore likely functional. A secondary structural model for small-subunit (18S) rRNA of Rafflesia keithii was shown in Nickrent and Starr (1994).
These results presented a number of challenges to concepts held about accumulation of mutations. The observation that mutational differences (amino acid level) in cytochrome c varied according to time of divergence of the organisms spawned the concept of the molecular clock (Zuckerkandl and Pauling 1965). A modification of this strict molecular clock idea derived from neutral theory predicts that rates will increase with shorter generation time. This concept was supported by molecular evolutionary studies of rodents and humans (Wu and Li 1985) as well as plants such as palms and grasses (Gaut et al. 1992). Our work with 18S rDNA in parasitic plants such as Rafflesia, Rhizanthes and Balanophora shows that substitution rates are inconsistent with a generation-time effect (Nickrent and Starr 1994). For example, Rafflesia has one of the highest substitution rates, much higher than Arabidopsis, yet its life cycle is much more protracted. This indicates that other factors, possibly population dynamics and molecular drive, are influencing substitution rates. Until recently, most work on 18S rDNA in plants was on crop species that do not show high evolutionary rates. In fact, the number of mutational differences between genera of most angiosperm families is too low to allow adequate resolution using this molecule. In contrast, 18S rDNA in the santalalean holoparasites is so divergent that this molecule cannot be used in global comparisons with other flowering plants because of long branch attraction problems (Nickrent and Starr 1994). The phylogenetic placement of these unusual families must wait until a suitable conservative molecular marker can be identified and applied to this problem.
Aside from the work on nuclear rDNA described above, the majority of molecular studies on parasitic plants has involved plastid DNA (ptDNA). A great deal of information on the plastid genome has been obtained from Cuscuta (Machado and Zetsche 1990, Haberhausen and Zetsche 1994, ) and Epifagus and Conopholis, two members of the broomrape family (Orobanchaceae). The complete plastid DNA sequence for Epifagus virginiana (beechdrops) has been determined (Wolf et al. 1992) which showed it to be only 71 kb in length as compared with tobacco which is 156 kb. Most of the deletions involve genes associated with photosynthesis (dePamphilis and Palmer 1990). The cpDNA genome of Conopholis americana (squawroot) has gone the next step further in reduction and has a cp genome that contains only one inverted repeat, hence its genome is estimated to be only 43.35 kb in length (Colwell 1994).
The purpose of our research on the ptDNA of the santalalean holoparasites was to first determine whether these plants have retained ptDNA at all. The first positive evidence for this came from our PCR experiments on 16S rDNA. Given that both Epifagus and Conopholis have a conserved inverted repeat containing the gene for 16S rRNA, we assumed that if any gene were still present in a plastid genome, it would be for ribosomal RNA. Since 16S rRNA is most like that of cyanobacteria we obtained sequences from Genbank, conducted an alignment containing cyanobacteria (e.g. Anacystis, Anabaena), green plants (Marchantia, Pinus, Nicotiana, etc.) and holoparasites, and identified regions on conservation. Oligonucleotide primers conserved between cyanobacteria and plant plastid genomes were obtained from Carl Woese (University of Illinois) and additional primers were designed based upon conserved sequences found in the above higher plants.
Results of our PCR amplifications from the three santalalean holoparasite lineages have shown that 16S rDNA exists in all representatives tested. Complete or nearly complete sequences have been obtained for Cynomorium (Balanophoraceae), Hydnora and Prosopanche (Hydnoraceae), Cytinus and Mitrastema (Rafflesiaceae s. lat.). For the latter family, partial sequences have also been obtained for Rafflesia, Pilostyles and Apodanthes. By making pairwise comparisons from aligned sequences, the number of nucleotide substitutions (transitions and transversions) between the two sequences can be tabulated. Published 16S rDNA sequences and those of the holoparasites were compared with tobacco (Nicotiana) and the results are shown in Figure 3. It can be seen that comparing a distantly related liverwort (Marchantia) with tobacco results in ca. 50 total substitutions. This number differs little from that obtained when tobacco is compared with another dicot such as Pisum (48 changes). The comparisons involving the santalalean holoparasites, however, showed a markedly greater number of substitutions. In the case of Hydnora, there are five-fold more substitutions than are observed in the liverwort-tobacco comparison. These results demonstrate that these holoparasites possess the most highly divergent plastid 16S rDNA sequences obtained to date.
Given the large number of changes in the holoparasite sequences, it is possible that they are nonfunctional pseudogenes. Secondary structure models of the 16S ribosomal RNAs (rRNAs) were constructed for all holoparasites sequenced to date. These models correspond well to previously proposed ones for cyanobacteria and green plant plastid rRNAs as evidenced by conservation of all major structural features (Gutell et al. 1985). Examination of the types of mutations occurring on the rRNA show that, despite frequent mutations, the majority do not disrupt secondary structure, i.e. they occur as compensated changes or in unpaired (loop) regions. The conclusion from this exercise is that the 16S rRNAs in these plants are very likely functional.
The conclusions from our work to date are as follows. A 16S rRNA gene that is homologous to other green plant 16S rRNAs has been amplified from representatives of three holoparasite families: Balanophoraceae, Hydnoraceae, and Rafflesiaceae. When compared with other angiosperms, these holoparasites show a very high number of nucleotide substitutions, higher than pairwise comparisons involving taxa known to be phylogenetically more distant. Despite the high rate of change, these rRNA sequences can be folded into a secondary structure therefore suggesting functionality. Our continuing work is aimed at further characterizing the chloroplast genome by PCR-based mapping analyses. Our preliminary results show that the 23S rDNA is present as well as several ribosomal proteins, thus supporting the concept that the plastid genome has been retained in these plants.
When molecular evidence is presented within the context of conservation biology, it usually involves conservation genetics. This rapidly expanding and increasingly relevant discipline has embraced the use of molecular methods to provide baseline populational genetic data on rare and endangered species. These data are used primarily to inform decisions on the preservation and management of populations experiencing loss of genetic diversity. Many of the parasitic plants that have been the subject of this paper certainly fall into this category. The "charismatic megaflora" such as Rafflesia are obvious candidates for in situ preservation given their requirement of mature, undisturbed forest ecosystems, curious biology, appeal to the ecotourist, etc. Other members of this group, such as the more cryptic and less charismatic Mitrastema, however, may not attract as much attention as Rafflesia and may, therefore, be overlooked as primary candidates for conservation.
By their very nature, parasitic plants, such as mistletoes, have complex associations with other organisms and are therefore more vulnerable than autotrophs to the effects of habitat perturbation. It is not clear how the elimination of parasitic species will impact the overall stability of the ecosystem. For example, the seeds of many Loranthaceae are specifically dispersed by flowerpeckers of the family Dicaeidae (Liddy 1983). Honeyeaters and sunbirds are frequent pollinators while foraging nectar from mistletoe flowers. Finally, the leaves of Loranthaceae in the 'Amylothecae' group are fed upon by butterfly caterpillars of the genus Delias (family Pieridae). It is also likely that complex associations with other organisms (e.g. fungi) are required for seed germination and seedling establishment for many holoparasites. Unfortunately, for the vast majority of the parasitic plants shown in Table 1, information is incomplete about basic life cycle parameters.
Nearly one quarter of the genera of Loranthaceae occur in Malaysia and some, such as Lampas, are found nowhere else (Borneo). The large, red, bird-pollinated flowers of Lepeostegeres are borne in capitate inflorescences and provide as striking a floral display as any tropical plant. Three species of this mistletoe are endemic to Borneo and should be considered candidates for conservation. Comparisons of localities of historical collections taken from southern Sarawak with current areas undergoing rapid urban development indicate that these populations are no longer in existence. Similarly, many of the mistletoe localities documented by collections made by Clemens in the 1930's from lower elevations of Mt. Kinabalu have been heavily impacted and likely do not currently support the required host tree species.
The application of molecular methods to parasitic plants goes beyond simply documenting levels of variation. These plants have undergone tremendous changes at the molecular level unknown in any other plant. For this reason, they should be viewed as model organisms that provide unprecedented opportunity for studying evolutionary change. The difficulty in placing Rafflesiaceae within the overall classification of angiosperms can be attributed to their extremely reduced vegetative body combined with novel floral morphological features. The molecular evidence obtained to date for these plants predicts that the genes involved in the development of these organs are also undergoing novel changes at the biochemical level. In his treatise on plant speciation in Malesia, Van Steenis (1969) also wondered about the evolutionary mechanisms that have produced such plants as Nepenthes, Rafflesia, Rhizanthes and Mitrastema. He proposed a saltatory model whereby novel innovations appear rapidly from symbiotic associations (in the case of parasites and saprophytes) and morphological and anatomical abnormalities (terata).
The reorganization of ptDNA in the holoparasites represents a natural genetic experiment that, upon careful study, can allow a more complete understanding of the structure and function of this genome in all plants. Since the phenomena we have observed at the molecular level are pervasive, occurring in unrelated plants from distant parts of the world, a number of questions are raised. What are the molecular mechanisms that are responsible for such drastic modification of the plastid genome? Are the same mechanisms operative in all organisms that evolve specialized nutritional dependence? If so, how does that impact upon the concept of a strict molecular clock? What are the functions of the genes that are retained in a vestigial plastid genome in a nonphotosynthetic plant? How many nuclear genes and gene products are utilized by the chloroplast and how did this symbiosis come about? One could ask, "why is it important to have answers to such apparently academic questions?" These questions would never have been asked if plants such as Balanophora, Hydnora, Rafflesia or Mitrastema were not available for study. In fact, it will only be through the study of these "exceptional" organisms that progress will be made in understanding the mode and tempo of evolutionary change at the molecular level. For example, the rRNA molecules of these plants have undergone amazingly high mutation pressure, yet are still functional. Study of the structure of these molecules can provide general insight into how all rRNA molecules function.
I began this paper with a general rationale for conserving biodiversity and then focused the discussion towards our molecular work on parasitic flowering plants. As a scientist, the "amenity value" I place on these plants stems from curiosity and a overall intrigue about their evolution, molecular and morphological specializations, and complex interactions with other organisms. Enough questions remain to fill many lifetimes of research - a legacy we should feel obligated to leave for future generations of biologists. I cannot claim that this justification is not anthropocentric, only that conserving such "curiosities" is the hallmark of an advanced civilization.
I wish to thank Joel Duff, Wei-xing Zong, Ellen Starr, Jamie Beam, and Erica Grimm for providing laboratory assistance with these "problem children" plants. I am grateful to Carl Woese (University of Illinois at Urbana/Champaign) for sending bacterial 16S rDNA primers that allowed our first PCR amplification of a plastid gene in a santalalean holoparasite. Sincere thanks to the following people who collected some of the parasite species shown in Figure 2. Herbarium voucher specimens are deposited at SIU (Nickrent accession number follows name): C. Augspurger (Heisteria concinna - 2732), D. E. Bran (Misodendron brachystacheum - 2829), W. Forstreuter (Champereia manillana - 3014), S. Medbury (Strombosia philippinensis - 2831), M. Melampy (Dendrophthora clavata - 2182), B. Molloy (Tupeia antarctica - 2742), R. Narayana (Santalum album - 2730), J. Paxton (Phoradendron californicum - 2689), S. Sargent (Gaiadendron punctatum - 2729; Antidaphne viscoidea - 2730), K. Steiner (Moquinella rubra - 3042), and W. Takeuchi (Korthalsella latissima - 1975). In addition, for the parasitic plants shown in Fig. 3, Danny Joel (Cynomorium coccineum - 4000), Wilhelm Barthlott (Cytinus ruber - 2738), Willem Meijer (Mitrastema yamamotoi - 2941), Sherwin Carlquist (Hydnora africana - 2767).
Barlow, B. A. 1983. Biogeography of Loranthaceae and Viscaceae. Pp. 19-45 In: The Biology of Mistletoes, D. M. Calder and P. Bernhardt, eds. Academic Press, New York.
Barlow, B. A. 1991a. Provisional key to the genera of Loranthaceae and Viscaceae of the Flora Malesiana region. Flora Malesiana Bull. 10:335-338.
Barlow, B. A. 1991b. Conspectus of the genera Scurrula L. and Taxillus Teighem (Loranthaceae) in the Malesian region. Blumea 36:63-85.
Barlow, B.A. 1993. Conspectus of the genera Amylotheca, Cyne, Decaisnina, Lampas, Lepeostegeres, and Loxanthera (Loranthaceae). Blumea 38:65-126.
Bhandari, N. N. and C. A. Vohra. 1983. Embryology and affinities of Viscaceae. Pp. 69-86 In: The Biology of Mistletoes, M. Calder and P. Bernhardt, eds. Acad. Press, New York.
Colwell, A. E. 1994. Genome evolution in a non-photosynthetic plant, Conopholis americana. Ph.D. Dissertation, Washington University, St. Louis, Missouri, USA. 222 pp.
Cronquist, A. 1988. The evolution and classification of flowering plants, 2nd. ed. The New York Botanical Garden, Bronx, NY.
dePamphilis, C. W. and J. D. Palmer. 1990. Loss of photosynthetic and chlororespiratory genes from the plastid genome of a parasitic flowering plant. Nature 348:337-339.
Ehrenfeld, D. 1988. Why put a value on biodiversity? Pp. 212-223 In: Biodiversity, E. O. Wilson, ed. National Acad. Press, Washington, D.C.
Gaut, B. S., S. V. Muse, W. D. Clark, and M. T. Clegg. 1992. Relative rates of nucleotide substitution at the rbcL locus of monocotyledonous plants. J. Mol. Evol. 35:292-303.
Gutell, R. R., B. Weiser, C. Woese, and H. F. Noller. 1985. Comparative anatomy of 16-S-like ribosomal RNA. Progr. Nucl. Acid Res. Mol. Biol. 32:155-216.
Grazi, G. 1984. Successful cultivation of pine-mistletoe (Viscum album subsp. austriacum) outside its natural area of distribution. Golden Bough 5:4-5.
Haberhausen, G. and K. Zetsche. 1994. Functional loss of all ndh genes in an otherwise relatively unaltered plastid genome of the holoparasitic flowering plant Cuscuta reflexa. Plant Mol. Biol. 24:217-222.
Hiepko, P. 1979. A revision of Opiliaceae I. Genera of the eastern Old World, excluding Opilia. Willdenowia 9:13-56.
Hiepko, P. 1982. A revision of Opiliaceae II. Opilia Roxb. Willdenowia 12:161-182.
Ismail, G. 1988. Conservation of the giant Rafflesia in Sabah, Malaysia. TREE 3:316-317.
Kuijt, J. 1969. The Biology of Parasitic Flowering Plants. Univ. California Press, Berkeley. 246 pp.
Kuijt, J. 1988. Monograph of Eremolepidaceae. Syst. Bot. Monog. 18:1-60.
Liddy, J. 1983. Dispersal of Australian mistletoes: the Cowiebank study. Pp. 101-116 In: The Biology of Mistletoes, D. M. Calder and P. Bernhardt, eds. Academic Press, New York.
Machado, M. A. and K. Zetsche. 1990. A structural, functional and molecular analysis of plastids of the holoparasites Cuscuta reflexa and Cuscuta europaea. Planta 181:91-96.
Meijer, W. 1984. New species of Rafflesia (Rafflesiaceae). Blumea 30: 209-215.
Meijer, W. 1985. Saving the world's largest flower. Natl. Geogr. Mag. 168: 136-140.
Meijer, W. Rafflesiaceae. In: Flora Malesiana, Rijksherbarium, Leiden, The Netherlands. (to be published 1996).
Meijer, W. and J. F. Veldkamp. 1988. A revision of Rhizanthes (Rafflesiaceae). Blumea 33: 329-242.
Nickrent, D. L. and C. R. Franchina. 1990. Phylogenetic relationships of the Santalales and relatives. J. Mol. Evol. 31:294-301.
Nickrent, D. L. and E. M. Starr. 1994. High rates of nucleotide substitution in nuclear small-subunit (18S) rDNA from holoparasitic flowering plants. J. Mol. Evol. 39:62-70.
Nickrent, D. L. and D. E. Soltis. 1995. A comparison of angiosperm phylogenies based upon complete 18S rDNA and rbcL sequences. Ann. Missouri Bot. Gard. 82:208-234.
Norton, B. 1988. Commodity, amenity, and morality. The limits of quantification in valuing biodiversity. Pp. 200-205 In: Biodiversity, E. O. Wilson, ed. National Acad. Press, Washington, D.C.
Norton, D. A. 1991. Trilepidea adamsii: an obituary for a species. Cons. Biol. 5:52-57.
Sleumer, H. 1980. A taxonomic account of the Olacaceae of Asia, Malesia, and the Adjacent Areas. Blumea 26:145-168.
Swofford, D. L. 1993. PAUP: phylogenetic analysis using parsimony, version 3.1. Smithsonian Institution, Washington, D.C.
Van Steenis, C. G. 1969. Plant speciation in Malesia, with special reference to the theory of non-adaptive saltatory evolution. Biol. J. Linn. Soc. 1:97-133.
Wiens, D. and B. A. Barlow. 1971. The cytogeography and relationships of the viscaceous and eremolepidaceous mistletoes. Taxon 20:313-332.
Wolf, K. H., C. W. Morden, et al. 1992. Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc. Nat. Acad. Sci. USA 89:10648-10652.
Wu, C.-I. and W.-H. Li. 1985. Evidence for higher rates of nucleotide substitution in rodents than in man. Proc. Natl. Acad. Sci. USA 82:1741-1745.
Zuckerkandl, E. and L. Pauling. 1965. Evolutionary divergence and convergence in proteins. Pp. 97-116 In: B. Bryson and H. J. Vogel, eds. Academic Press, New York.
Table 1. Santalalean Parasites of Malaysia
| Fam./Genus | Malaysian Species | Total Species | Fam./Genus | Malaysian Species | Total Species |
| Olacaceae | Santalaceae | ||||
| 1. Anacolosa | 1 | 16 | 1. Cladomyza | 1 | ca. 17 |
| 2. Erythropalum | 1 | 1 | 2. Dendrotrophe | 2? | 30 |
| 3. Harmandia | 1 | 1 | 3. Dufrenoya | 1? | ca. 14 |
| 4. Ochanostachys | 1 | 1 | 4. Exocarpos | 1 | 26 |
| 5. Olax | 2 | 40 | 5. Phacellaria | 1 | 7 |
| 6. Schoepfia | 1 | 30 | 6. Santalum | 1 | 25 |
| 7. Scorodocarpus | 1 | 1 | 7. Scleropyrum | 1 | 6 |
| 8. Strombosia | 2 | 16 | |||
| 9. Ximenia | 1 | 9 | Viscaceae | ||
| 1. Ginalloa | 3 | 5 | |||
| Opiliaceae 2 | 2. Korthalsella | 2 | ca. 30 | ||
| 1. Cansjera | 1 | 3 | 3. Notothixos | 1 | 8 |
| 2. Champereia | 1 | 1 | 4. Viscum | 4 | ca. 100 |
| 3. Lepionurus | 1 | 1 | |||
| 4. Melientha | 1 | 1 | Balanophoraceae | ||
| 5. Opilia | 1 | 3 | 1. Balanophora | 5 | 15 |
| 6. Urobotrya | 1 | 5 | 2. Exorhopala | 1 | 1 |
| 3. Rhopalocnemis | 1 | 1 | |||
| Loranthaceae 3 | |||||
| 1. Amyema | 4 | ca. 100 | Rafflesiaceae | ||
| 2. Amylotheca | 1 | 5 | 1. Mitrastema | 1 | 2 |
| 3. Barathranthus | 2 | 3 | 2. Rafflesia | 8 | 13 extant |
| 4. Dendrophthoe | ca. 5 | 30 | 3. Rhizanthes | 2 | 2 |
| 5. Elytranthe | 1 | 10 | |||
| 6. Helixanthera | ca. 10 | 50 | |||
| 7. Lampas 4 | 1 | 1 | |||
| 8. Lepeostegeres | 5 | 9 | |||
| 9. Lepidaria | 6 | 13 | |||
| 10. Loxanthera | 1 | 1 | |||
| 11. Macrosolen | ca. 13 | 25 | |||
| 12. Scurrula | 3 | 20 | |||
| 13. Taxillus | 1 | 30 | |||
| 14. Trithecanthera | 1 | 4 |
Table 2. The genus Rafflesia R. Brown 1
| Species | Distribution |
1. R. arnoldii R. Brown = R. titan Jack = R. tuan-mudae Beccari |
Sarawak, Sumatra, W. Kalimantan Sumatra Sarawak, Borneo |
| 2. R. borneensis Koorders | N. Borneo |
| 3. R. cantleyi Solms-Laubach | Peninsular Malaysian |
4. R. ciliata Koorders 2 = R. witkampii Koorders 2 |
N. Borneo N. Borneo |
| 5. R. gadutensis Meijer | Western coastal Sumatra |
| 6. R. hasseltii Suringar | Sumatra, peninsular Malaysia |
| 7. R. keithii Meijer | Sabah, Malaysia |
| 8. R. kerrii Meijer | Peninsular Thailand and Malaysia |
| 9. R. manillana Teschemacher | Philippines |
| 10. R. micropylora Meijer | Sumatra |
11. R. patma Blume = R. zollingeriana Koorders |
Java Java |
| 12. R. pricei Meijer | Sabah, Malaysia |
| 13. R. rochussenii Teijsm. & Binnend. | Western Java, Sumatra |
| 14. R. schadenbergiana Goeppert 2 | Philippines |
| 15. R. tengku-adlinii Salleh | Borneo |
Figure 1. Occurrence of parasitism in angiosperms mapped upon the subclass classification of Cronquist (1988). In that system, Balanophoraceae was placed in Santalales whereas Hydnoraceae and Rafflesiaceae were placed in the allied Rafflesiales. Molecular phylogenetic studies do not support this association but have not, as yet, revealed the correct placement of these families. Eremolepidaceae are here included in Santalaceae (see text).
Figure 2. Majority rule consensus phylogram of 10 equally most parsimonious trees of length 2262 resulting from heuristic analysis of complete 18S rDNA and rbcL sequences. Numbers above the branches indicate branch length (number of substitutions). Numbers below branches represent the percentage of trees (out of 10) supporting the clade. All nodes without numbers were found in all 10 trees.
Figure 3. Number of substitutions (transitions and transversions) resulting from pairwise comparisons of plastid-encoded 16S rDNA between Nicotiana and other green plants. The santalalean holoparasites (Cynomorium, Cytinus, Mitrastema and Hydnora) show a marked increase in number of substitutions.
SIUC / College of Science / Parasitic Plant Connection /
URL: http://www.parasiticplants.siu.edu/Malay.biodiv.html
Last updated: 6-Feb-99 / dln